Oncomedicine 2016; 1:14-17. doi:10.7150/oncm.16802 This volume Cite
Short Research Paper
Cathelicidin LL-37 Promotes or Inhibits Cancer Cell Stemness Depending on the Tumor Origin
1. Emergency Medicine Department, University of Sao Paulo, Sao Paulo, Brazil
2. Oncology Department, University of Sao Paulo, Sao Paulo, Brazil
Received 2016-7-20; Accepted 2016-8-15; Published 2016-8-21
Abstract
Antimicrobial peptides play critical protective roles in a range of human diseases, including cancer. Multiple studies have demonstrated functions—such as proliferation, angiogenesis, apoptosis and immunomodulation—of these peptides in crucial cancer pathways. We investigated the role of the antimicrobial peptide LL-37 on stemness in breast cancer (SKBR3) and melanoma cells (A375). PCR array analysis of differential gene expression in SKBR3 and A375 cancer cell lines downregulated for LL-37 expression by siRNA revealed downregulation of genes related to stemness, including telomerase reverse transcriptase, forkhead box D3 and undifferentiated embryonic cell transcription factor 1, remarkably in breast cancer cells. Furthermore, SKBR3 cells knocked down for LL-37 expression showed a decreased production of oncospheres in comparison with negative controls, while A375 cells exhibited increased production. Taken collectively, our findings indicate a role for LL-37 in cancer cell stemness depending on the cell type.
Keywords: LL-37, cancer, stemness, pluripotency, self-renewal
Introduction
Antimicrobial peptides play crucial roles in critical molecular pathways in cancer, such as cell proliferation, epithelial cell migration, angiogenesis promotion, induction of apoptosis and immunomodulation (1).
The effects of the antimicrobial peptide LL-37 in cancer remain unclear. While LL-37 acts as a positive regulator of ovarian, breast, melanoma and lung cancer progression, it also suppresses colorectal and gastric cancer cell growth (2), indicating that its effects are tumor-specific.
Recently, the concept of clonal tumor evolution has been challenged by the observation of cancer stem cells (CSCs) in a variety of tumors. CSCs possess increased invasive and metastatic capabilities and render tumors more resistant to several microenvironmental stresses, including the action of several anti-cancer drugs (3).
Here we investigated the effects of LL-37 on stemness in both breast cancer and melanoma cells. We performed array analysis to examine the expression of 84 genes related to DNA damage in wild-type and LL-37-knockdown cancer cells.
Material and Methods
The study protocol was approved by the Hospital das Clinicas Ethical Committee, protocol 034/14.
Cell culture
Immortalized human breast cancer cells (SKBR-3) and skin malignant melanoma cells (A375) were used in this study. Cells were maintained according to the guidelines of the ATCC (American Type Culture Collection).
Real-time PCR
RNA was extracted from cultured cells using TRIZOL® protocol. RNA was quantified using NanoVeu (GE Heathcare) systems and RT-PCR was performed using the StepOne SuperScript® III (Applied Biosystems) protocol as provided by the manufacturer. Beta-2 microglobulin (B2M) gene was used as an internal control; primers were as follows: GAT GAG TAT GCC TGC CGT TGC, and CAA TCC AAA TGC GGC ATC T. The reactions were performed in a StepOne™ system (Applied Biosystems) at 50°C for 10 min, 95°C for 5 min and then 95°C for 15 s followed by 60°C for 30 s, and 72°C for 30 s for 40 cycles. Quantification was performed by 2-ΔΔCT method.
LL-37 gene silencing
Cells were plated at 2.5×105 cells per well in a 6-well plate overnight. LL-37 Silencer Selected Pre-designed short interfering RNA (siRNA) or negative scramble siRNA (Ambion®) (10 nM each) was combined with 5 µL of Lipofectamine™ RNAiMAX reagent for 20 min. Opti-MEM® I Reduced Serum Medium (Invitrogen) was added to a final volume of 2.5 mL per well after cells were rinsed with PBS. After 24 h (SKBR3) or 48 h (A375), experimental assays were performed.
PCR array
Total RNA was converted into cDNA using the RT2 First Strand Kit (SABiosciences, Frederick, USA) and cDNA was then combined with the RT2 SYBR Green qPCR Master Mix (SABiosciences). Each sample was added to 24 Human DNA Damage PCR arrays (Qiagen, USA) according to the StepOne equipment protocol (Applied Biosystems). PCR-array data analysis was performed at the manufacturer's website (http://www.sabiosciences.com/pcrarraydataanalysis.php).
Sphere-forming assays
The assay was performed as previously reported (4, 5). Briefly, after siRNA treatment, cells were detached and a single cell suspension was obtained after passing cells through 25 G needles. Cells (1.0×105) were plated in their respective cell culture media containing B27 supplement and rEGF (100 ng/ml; Sigma Aldrich, Poole, UK; E-9644). After 5 days, the number of spheres that were greater than 50 µm in diameter were counted and sphere forming efficiency (%) was determined.
Statistical analysis
Results were analyzed suing Mann-Whitney test and are shown as mean ± standard deviation. A p-value < 0.05 was considered significant.
Results
LL-37 production upregulates several pathways related to stemness
We next analyzed differential gene expression in cancer cell lines downregulated for LL-37 expression by siRNA. Knockdown efficiency of LL-37 expression was more than 90%, as evaluated by qPCR (data not shown). PCR arrays showed a downregulation of several genes related to stemness, especially in SKBR3, for LL-37 compared with control cells (Tables 1 and 2).
LL-37 knockdown cells show decreased production of oncospheres in breast cancer cells and increased production in melanoma cells
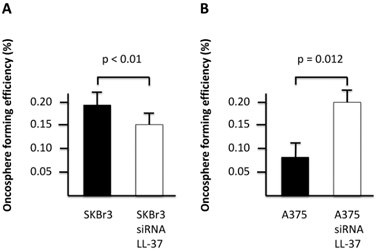
We next analyzed the production of cancer-derived extracellular vesicles, oncospheres, as a hallmark of stemness in cancer cell lines depleted for LL-37 expression. Our results showed that SKBR3 cells knocked down for LL-37 expression produced a decreased number of oncospheres compared with negative controls (Figure 1A), while A375 produced an increased number of oncospheres compared with negative controls (Figure 1B).
Oncosphere-forming efficiency in SKBR3 (A) and A375 (B) cell lines depleted for LL-37 expression by siRNA.
Genes upregulated in SKBR3 breast cancer cells compared with SKBR3 cells transfected with LL-37 siRNA treatment. Genes related to stemness are in bold.
| Symbol | Gene Name | Fold Change | p-value |
|---|---|---|---|
| KAT2A | K(lysine) acetyltransferase 2A | 1.6337 | 0.008484 |
| COL2A1 | Collagen, type II, alpha 1 | 1.8903 | 0.012853 |
| GDF3 | Growth differentiation factor 3 | 1.8402 | 0.014975 |
| HNF4A | Hepatocyte nuclear factor 4, alpha | 1.9064 | 0.016682 |
| TERT | Telomerase reverse transcriptase | 2.2468 | 0.018056 |
| HAND1 | Heart and neural crest derivatives expressed 1 | 2.1076 | 0.02039 |
| HSPA9 | Heat shock 70kDa protein 9 (mortalin) | 3.7997 | 0.020459 |
| SOX15 | SRY (sex determining region Y)-box 15 | 2.1014 | 0.02309 |
| NAT1 | N-acetyltransferase 1 (arylamine N-acetyltransferase) | 26.925 | 0.026202 |
| ALPL | Alkaline phosphatase, liver/bone/kidney | 1.838 | 0.026346 |
| PARD6A | Par-6 partitioning defective 6 homolog alpha (C. elegans) | 14.2326 | 0.026805 |
| OLIG2 | Oligodendrocyte lineage transcription factor 2 | 1.7598 | 0.031648 |
| FOXD3 | Forkhead box D3 | 2.6502 | 0.03305 |
| MYBL2 | V-myb myeloblastosis viral oncogene homolog (avian)-like 2 | 1.6863 | 0.035958 |
| LIN28A | Lin-28 homolog A (C. elegans) | 1.8108 | 0.036787 |
| TP53 | Tumor protein p53 | 1.9757 | 0.040021 |
| FGF2 | Fibroblast growth factor 2 (basic) | 2.0355 | 0.044985 |
| NES | Nestin | 1.8498 | 0.047692 |
| UTF1 | Undifferentiated embryonic cell transcription factor 1 | 5.2139 | 0.048209 |
Discussion
Self-renewal and pluripotency are the key characteristics of stem cells. Here, our findings suggest that LL-37 regulates stemness. Cancer stem cells (CSCs) are a considerable clinical problem, since they are highly resistant to radiation and chemotherapy (6). The mechanism of resistance is not fully understood, but enhanced DNA repair capacities and low intracellular reactive oxidative species concentrations are implicated (7). CSCs also proliferate more slowly than non-stem carcinoma cells, circulate in the bloodstream (8) and can lead to tumor metastasis and relapse. Definitive markers of CSCs do not exist, but our results demonstrated many interesting results in SKBR3 and A375 wild-type cells, when compared with LL-37 knockdown cells. Nestin, for example, is an important marker of CSCs, regulates proliferation, migration and invasion of cancer cells (9) correlating to a worse prognosis (10).
Telomerase reverse transcriptase, forkhead box D3 (FOXD3) and undifferentiated embryonic cell transcription factor 1 (UTF1) are other genes related to stemness, so we hypothesized that LL-37 should be important to maintain stem cell identity in breast cancer cells (Table 1). Upregulation of telomerase is a prerequisite for cellular immortalization and has been associated with stemness in various human cancers (11). FOXD3 induces cancer progression by epithelial-mesenchymal transition (12) and UTF1 increases stem cells reprogramming to pluripotency (13).
Genes upregulated in A375 melanoma cells compared with A375 cells transfected with LL-37 siRNA treatment. Genes related to stemness are in bold.
| Symbol | Gene Name | Fold Change | p-value |
|---|---|---|---|
| LIN28A | Lin-28 homolog A (C. elegans) | 1.5252 | 0 |
| TCF3 | Transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | 2.9868 | 0 |
| RUNX2 | Runt-related transcription factor 2 | 1.7998 | 0.000007 |
| CCNA2 | Cyclin A2 | 1.3786 | 0.000011 |
| FOXA2 | Forkhead box A2 | 1.5997 | 0.000018 |
| CD34 | CD34 molecule | 1.3355 | 0.000025 |
| BGLAP | Bone gamma-carboxyglutamate (gla) protein | 1.5147 | 0.000044 |
| CDH2 | Cadherin 2, type 1, N-cadherin (neuronal) | 1.6844 | 0.000044 |
| FABP7 | Fatty acid binding protein 7, brain | 1.6157 | 0.000056 |
| CCNE1 | Cyclin E1 | 1.472 | 0.000063 |
| OLIG2 | Oligodendrocyte lineage transcription factor 2 | 1.5713 | 0.000086 |
| NCAM1 | Neural cell adhesion molecule 1 | 1.2164 | 0.00013 |
| REST | RE1-silencing transcription factor | 1.3996 | 0.000231 |
| TUBB3 | Tubulin, beta 3 | 1.4953 | 0.000248 |
| CDC42 | Cell division cycle 42 (GTP binding protein, 25kDa) | 1.5889 | 0.00025 |
| KRT15 | Keratin 15 | 1.469 | 0.000278 |
| GATA2 | GATA binding protein 2 | 1.5547 | 0.000372 |
| ALPL | Alkaline phosphatase, liver/bone/kidney | 1.4816 | 0.000379 |
| GDF3 | Growth differentiation factor 3 | 1.7102 | 0.000422 |
| TBX3 | T-box 3 | 1.5775 | 0.000669 |
| AICDA | Activation-induced cytidine deaminase | 1.3863 | 0.001057 |
| RUNX1 | Runt-related transcription factor 1 | 1.2238 | 0.001949 |
| KLF4 | Kruppel-like factor 4 (gut) | 1.1139 | 0.00216 |
| FGF2 | Fibroblast growth factor 2 (basic) | 1.4219 | 0.002267 |
| NAT1 | N-acetyltransferase 1 (arylamine N-acetyltransferase) | 1.3888 | 0.002528 |
| LEFTY2 | Left-right determination factor 2 | 1.3489 | 0.003161 |
| MYCN | V-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) | 1.1672 | 0.003271 |
| HAND1 | Heart and neural crest derivatives expressed 1 | 1.2117 | 0.003824 |
| ZFP42 | Zinc finger protein 42 homolog (mouse) | 1.2421 | 0.004222 |
| HSPA9 | Heat shock 70kDa protein 9 (mortalin) | 1.4029 | 0.005582 |
| FGFR1 | Fibroblast growth factor receptor 1 | 1.3626 | 0.005687 |
| FGF4 | Fibroblast growth factor 4 | 1.1789 | 0.006383 |
| NODAL | Nodal homolog (mouse) | 1.6944 | 0.0064 |
| ALDH1A1 | Aldehyde dehydrogenase 1 family, member A1 | 1.2544 | 0.007374 |
| KAT2A | K(lysine) acetyltransferase 2A | 1.2006 | 0.007585 |
| PAX6 | Paired box 6 | 1.1578 | 0.008689 |
| DNMT3B | DNA (cytosine-5-)-methyltransferase 3 beta | 1.2005 | 0.011056 |
| HNF4A | Hepatocyte nuclear factor 4, alpha | 1.4146 | 0.011275 |
| CDK1 | Cyclin-dependent kinase 1 | 1.1471 | 0.012381 |
| COL1A1 | Collagen, type I, alpha 1 | 1.1761 | 0.015877 |
| LEFTY1 | Left-right determination factor 1 | 1.226 | 0.015976 |
| DPPA3 | Developmental pluripotency associated 2 | 1.3138 | 0.024156 |
| NR5A2 | Nuclear receptor subfamily 5, group A, member 2 | 1.1412 | 0.026799 |
| EP300 | E1A binding protein p300 | 1.3847 | 0.029084 |
| KAT7 | K(lysine) acetyltransferase 7 | 1.967 | 0.029885 |
| PARD6A | Par-6 partitioning defective 6 homolog alpha (C. elegans) | 1.2703 | 0.033506 |
| POU5F1 | POU class 5 homeobox 1 | 1.1131 | 0.042862 |
| ESRRB | Estrogen-related receptor beta | 1.2021 | 0.044154 |
The results were not so evident in A375 melanoma cells, but the presence of Runt-related transcription factors 1 and 2 (14-16), Cadherin 2 (17, 18), Nodal homolog (mouse) (19) and Aldehyde dehydrogenase 1 (family member A1) pointed to the same direction (20) (Table 2). However, the production of oncospheres in A375 cells put in evidence that LL-37 has opposing effects on cancer cell stemness depending on the cell type.
Conclusion
Emerging evidence highlights the role of antimicrobial peptides in non-infectious diseases, such as cancer. The mechanisms triggered by antimicrobial peptides are broad and lead to unexpected cell responses. Here, we show that the antimicrobial peptide LL-37 is implicated in cancer stemness. Further research needs to be directed to better clarify this phenomenon and the role of other antimicrobial peptides in this scenario.
Acknowledgements
This work was supported by FAPESP, the Sao Paulo Research Foundation (grant # 2015/00892-4).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Piktel E, Niemirowicz K, Wnorowska U, Watek M, Wollny T, Gluszek K. et al. The Role of Cathelicidin LL-37 in Cancer Development. Archivum immunologiae et therapiae experimentalis. 2016;64(1):33-46
2. Pinheiro da Silva F, Machado MC. Antimicrobial peptides: clinical relevance and therapeutic implications. Peptides. 2012;36(2):308-14
3. Cojoc M, Mabert K, Muders MH, Dubrovska A. A role for cancer stem cells in therapy resistance: cellular and molecular mechanisms. Seminars in cancer biology. 2015;31:16-27
4. Shaw FL, Harrison H, Spence K, Ablett MP, Simoes BM, Farnie G. et al. A detailed mammosphere assay protocol for the quantification of breast stem cell activity. Journal of mammary gland biology and neoplasia. 2012;17(2):111-7
5. Sadovska L, Eglitis J, Line A. Extracellular Vesicles as Biomarkers and Therapeutic Targets in Breast Cancer. Anticancer research. 2015;35(12):6379-90
6. Butof R, Dubrovska A, Baumann M. Clinical perspectives of cancer stem cell research in radiation oncology. Radiotherapy and oncology: journal of the European Society for Therapeutic Radiology and Oncology. 2013;108(3):388-96
7. Skvortsova I, Debbage P, Kumar V, Skvortsov S. Radiation resistance: Cancer stem cells (CSCs) and their enigmatic pro-survival signaling. Seminars in cancer biology. 2015;35:39-44
8. Yang MH, Imrali A, Heeschen C. Circulating cancer stem cells: the importance to select. Chinese journal of cancer research = Chung-kuo yen cheng yen chiu. 2015;27(5):437-49
9. Narita K, Matsuda Y, Seike M, Naito Z, Gemma A, Ishiwata T. Nestin regulates proliferation, migration, invasion and stemness of lung adenocarcinoma. International journal of oncology. 2014;44(4):1118-30
10. Ishiwata T, Matsuda Y, Naito Z. Nestin in gastrointestinal and other cancers: effects on cells and tumor angiogenesis. World journal of gastroenterology. 2011;17(4):409-18
11. Terali K, Yilmazer A. New surprises from an old favourite: The emergence of telomerase as a key player in the regulation of cancer stemness. Biochimie. 2016;121:170-8
12. Chu TL, Zhao HM, Li Y, Chen AX, Sun X, Ge J. FoxD3 deficiency promotes breast cancer progression by induction of epithelial-mesenchymal transition. Biochemical and biophysical research communications. 2014;446(2):580-4
13. Galonska C, Smith ZD, Meissner A. In Vivo and in vitro dynamics of undifferentiated embryonic cell transcription factor 1. Stem cell reports. 2014;2(3):245-52
14. Samokhvalov IM. A long way to stemness. Cell cycle. 2012;11(16):2965-6
15. Niu DF, Kondo T, Nakazawa T, Oishi N, Kawasaki T, Mochizuki K. et al. Transcription factor Runx2 is a regulator of epithelial-mesenchymal transition and invasion in thyroid carcinomas. Laboratory investigation; a journal of technical methods and pathology. 2012;92(8):1181-90
16. Vasuri F, Resta L, Fittipaldi S, Malvi D, Pasquinelli G. RUNX-1 and CD44 as markers of resident stem cell derivation in undifferentiated intimal sarcoma of pulmonary artery. Histopathology. 2012;61(4):737-43
17. Pieters T, van Roy F. Role of cell-cell adhesion complexes in embryonic stem cell biology. Journal of cell science. 2014;127(Pt 12):2603-13
18. Farahani E, Patra HK, Jangamreddy JR, Rashedi I, Kawalec M, Rao Pariti RK. et al. Cell adhesion molecules and their relation to (cancer) cell stemness. Carcinogenesis. 2014;35(4):747-59
19. Stefanidis K, Pergialiotis V, Christakis D, Loutradis D, Antsaklis A. Nodal, Nanog, DAZL and SMAD gene expression in human amniotic fluid stem cells. Journal of stem cells. 2013;8(1):17-23
20. Khorrami S, Zavaran Hosseini A, Mowla SJ, Malekzadeh R. Verification of ALDH Activity as a Biomarker in Colon Cancer Stem Cells-Derived HT-29 Cell Line. Iranian journal of cancer prevention. 2015;8(5):e3446
Author contact
![]() Corresponding author: Fabiano Pinheiro da Silva, M.D., Ph.D. Faculdade de Medicina da Universidade de São Paulo, Laboratório de Emergências Clínicas (LIM-51), Av. Dr. Arnaldo, 455 sala 3189, CEP 01246-000, São Paulo - SP, Brazil. Phone: +55 11 3061 8480; Fax: +55 11 3061 8480; E-mail: pinheirofabianocom
Corresponding author: Fabiano Pinheiro da Silva, M.D., Ph.D. Faculdade de Medicina da Universidade de São Paulo, Laboratório de Emergências Clínicas (LIM-51), Av. Dr. Arnaldo, 455 sala 3189, CEP 01246-000, São Paulo - SP, Brazil. Phone: +55 11 3061 8480; Fax: +55 11 3061 8480; E-mail: pinheirofabianocom