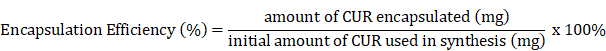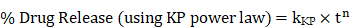Oncomedicine 2018; 3:94-109. doi:10.7150/oncm.27938 This volume Cite
Research Paper
Liposomes Aid Curcumin's Combat with Cancer in a Breast Tumor Model
1. Biomedical Engineering and Biotechnology Program, University of Massachusetts Lowell, 1 University Ave, Lowell, MA 01854
2. Department of Chemical Engineering, University of Massachusetts Lowell, 1 University Ave, Lowell, MA 01854.
* Authors contributed equally to this research article
Received 2018-6-17; Accepted 2018-9-21; Published 2018-10-5
Abstract
Curcumin (CUR), a natural compound, has multiple antineoplastic properties and specificity toward tumor cells, however its bioavailability and water solubility are poor. Liposomes increase the therapeutic index of CUR by protecting the drug from enzymatic degradation and can be made long circulating by surface modification with polyethylene glycol (PEG). Folate receptor α (FRα) is overexpressed in several types of cancer. Therefore, a folate-conjugated liposomal CUR nanoconstruct can enhance tumor targeted drug therapy. In this study, we synthesized, characterized and tested the cytotoxicity of CUR loaded liposomes (folate modified and non-folate modified) and CUR free drug in non-malignant breast epithelial cells and breast cancer cells, in addition to assessing the effects of CUR on cell cycle. The breast cancer cells not only had increased uptake of liposomes compared to non-malignant cells, but also more showed greater cytotoxicity resulted from treatments with free CUR and liposomal CUR (folate surface-modified and non-folate liposome types). The imaging and killing results, indicated that the liposomal CUR formulations do not alter the therapeutic efficacy and selectivity of CUR to provide anticancer benefits. Moreover, the differences in cellular uptake between the two liposomal nanoconstructs were studied using flow cytometry which showed targeting of folate surface-modified liposomes in cancer cells based on the findings presented.
Keywords: Drug delivery, oncology, nanomedicine, targeted therapy, nanotechnology, chemoprevention
Introduction
Curcumin (CUR) is a naturally derived phenolic compound from the rhizome Curcuma longa [1-4]. CUR is a small molecule hydrophobic drug derived from turmeric, a routinely consumed spice by several Asian communities. CUR has biological and pharmacological relevance making it an exciting molecule with anti-inflammatory, anti-carcinogenic, anti-malarial and anti-viral properties, in addition to low toxicity [3, 6-11]. Efforts by others have shown the selectivity of CUR towards tumor cells compared to healthy cells which embarked the use of curcumin for cancer treatment [5, 12-18]. CUR has multiple anticancer facets including blocking tumor initiation, as well as suppressing tumor progression and inhibiting invasion and metastasis [8, 12, 18-23]. CUR acts on matrix metalloproteinases, cyclooxygenase, vascular endothelial growth factor (VEGF), tumor necrosis factor alpha (TNF-α), among other signal transduction pathways [8, 12-15, 24, 25]. However, CUR as a free drug has a short plasma half-life because the liver rapidly metabolizes it [6]. Further, CUR is poorly water-soluble, <0.125 mg/L, and susceptible to enzymatic degradation in the blood stream, all of which limit the drug's bioavailability and therapeutic potential as a chemotherapeutic drug [5, 6, 15, 21, 24, 26, 27]. These issues can be addressed using drug delivery systems (DDS) to increase blood plasma residence time [28, 29]. Different forms of DDS have been explored to encapsulate curcumin for treatment of various cancer types [16, 17, 30-32].
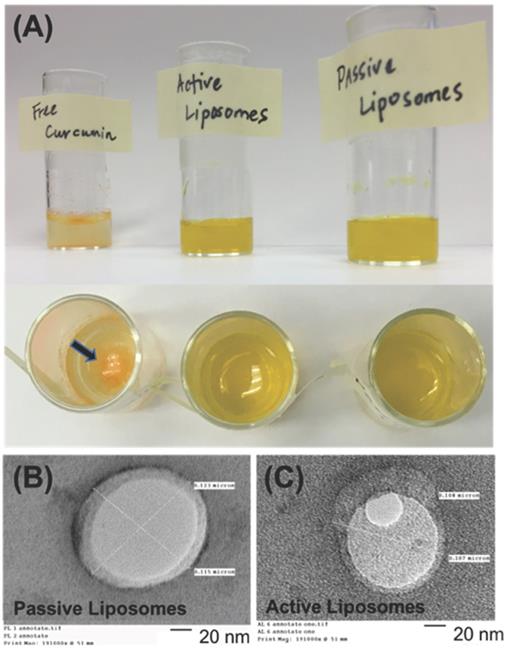
(A) Free CUR in water, active (folate conjugated) CUR liposomes, and passive CUR liposomes are shown to demonstrate how the free drug precipitates out of solution due to its poor water solubility, while liposomal formulations encapsulate CUR for improved drug delivery. A black arrow indicates the location of dark orange CUR precipitate in water. A previous study found the aqueous solubility of CUR to be <0.125 mg/L [5]. Transmission electron microscopy (TEM) images are shown in (B) and (C) for passive CUR liposomes and active CUR liposomes, respectively. TEM is used to confirm the sizes of liposomes, as well as the shape of the nanoconstructs. Scale Bar=20 nm.
In particular, liposomal nanoconstructs are extensively studied to improve the bioavailability of CUR, as these DDS are approved by the Food and Drug Administration (FDA) [3, 23, 33-37] [28, 38]. In fact, a study by Mondal et al demonstrated how effective liposomal encapsulation of CUR can be by increasing the solubility to 500 ug/mL, an impressive improvement compared to the aforementioned solubility value of <0.125 mg/L reported by Feng et al [5, 39]. The benefits of using a phospholipid bilayer nanoconstruct, as shown in Figures 1 (A), 1 (B), and 1 (C), include dual loading of hydrophilic and hydrophobic drugs, improved bioavailability to target site cell and tissue, stability of encapsulated drugs, pharmacological inactivity and minimal toxicity of phospholipids [29, 40]. Several investigations have shown the benefits of encapsulating curcumin in different cancer types such as pancreatic adenocarcinoma, osteosarcoma, liver cancer etc [41-44]. Different types of lipid based nanosystems, such as long circulating PEGylated, cationic liposomes and so on, have shown promise due to the flexibility of altering its properties by changing the composition and concentration of lipids [41, 45, 46]. Mahmud et. al. for example, demonstrated the anticancer activity of long-circulating liposomes encapsulating curcumin in vitro in pancreatic cancer cell lines. Kitajima et. al. demonstrated the anticancer activity of liposomal formulations of curcumin in vivo against osteosarcoma in mice [42]. Nanoemulsions encapsulating curcumin have also been explored for activity against cancer [43]. Several studies that have explored the use of nanoparticles for curcumin delivery are reviewed by Yallapu et. al. [10, 44]. Most of the studies involve the use of non-targeted, nano-formulations of curucmin [10, 44]. Enhanced delivery and uptake of curcumin encapsulated in liposomes was explored by Miyazawa et. al. by incorporating amadori-glycated phosphatidylethanolamine in the lipid vesicles which helped lower the negative membrane potential of the liposomes which improved cellular uptake [47]. Liposomal delivery of CUR has also been tested in clinical trials for cancer therapy [48]. Storka et al. found that intravenous doses up to 120 mg/m2 are safe short-term in healthy individuals [48]. In an ongoing phase 1b dose escalation study by SignPath Pharma, Inc., solid tumor cancer patients were given intravenous liposomal curcumin to determine the maximum tolerated dose (MTD), safety, and pharmacokinetic (PK) profiles of CUR [48]. No limiting adverse side effects have been reported thus far and the company is planning a Phase 2 trial in glioblastoma patients this year. Nutraceutical development of micronized CUR is also on the market as Source Naturals Theracurmin® which has 27 times more bioavailability than pure curcumin by blood plasma concentration [7, 18, 49, 50]. Theracurmin® is made with gum ghatti to improve gut absorption and enhance the effects of CUR in the body which range from cardiovascular health to free radical protection [7, 18, 50]. Other products such as Longvida® use solid lipid curcumin particle (SLCP) technology to enhance the bioavailability of CUR by protecting CUR from degradation in the stomach and promoting free drug absorption in the gut.
While extensive work has been conducted with the liposomal CUR formulation, our study focused on aiding the selectivity of CUR by actively targeting the liposomes to the overexpressed receptors on cancer cells. This was achieved by surface modification of liposomes with folic acid which binds to overexpressed folate receptors (FRα) on tumor cells [51-56]. FRα, a glycosylphosphatidylinositol (GPI) anchored cell surface protein, belongs to the folate receptor family which uses unidirectional transport of folate into cells [52, 56, 57]. FRα is found on the acinar cells of the breast [57]. The high affinity of folate receptors such as FRα for folate and folate analogs make this receptor an ideal target for folate conjugated liposomes [57]. These conjugates have been shown to have high affinity and specificity for folate receptors [52, 53]. The hypothesis of our study was that curcumin encapsulating folate targeted liposomes show preferential cellular uptake and killing of tumor cells compared to normal cells, as found in previous studies which used free CUR [6, 58] and to establish targeting potential of folate functionalized liposomes using flow cytometry experiment. To our knowledge, a direct comparative results showing the difference in curcumin efficiency between non-malignant and cancer cells using folate decorated liposomes has not been demonstrated thus far. To test this comparison in vitro and challenge the selective attribute of curcumin when encapsulated in a targeted liposomal modality, we decided to conduct a parallel study using non-malignant mammary epithelial cells, MCF-12A, and triple negative breast cancer cells (TNBC), MDA-MB-231[51, 59-63].
TNBC is the most aggressive form of breast cancer that accounts for approximately 15-20% of breast cancer cases in the U.S. [28, 64, 65]. TNBC cells do not express estrogen receptor (ER), progesterone receptor (PR), or human epidermal growth factor (HER-2) genes, rendering this type of cancer particularly difficult to treat due to a lack of endocrine response [65, 66]. Fortunately, TNBC shows sensitivity towards chemotherapy [67], making it viable for curcumin therapy. Furthermore, FRα is overexpressed on TNBC cells [51, 53, 68], providing an additional route for targeted treatment which can enhance cellular uptake of folate liposomes. Thus, the idea behind comparing cell lines was to demonstrate that CUR has preferential uptake and killing in MDA-MB-231, while remaining less harmful to MCF-12A.
We have synthesized, characterized and tested the cytotoxicity of CUR loaded liposomes and CUR free drug in normal breast cells and TNBC cells. We have also compared the cellular uptake of fluorescently labeled active (folate modified) empty liposomes and passive (non-folate modified) empty liposomes in normal and TNBC cells. This research was designed to provide a foundation for understanding the comparative effects of liposomal curcumin on normal breast epithelial cells and TNBC cells. While curcumin nanoformulations have been tested in the treatment of breast cancer, our study is unique in that, to the best of our knowledge, no other research has investigated folate-targeted CUR liposomes in MCF-12A and MDA-MB-231 cell lines or compared the cell lines directly for cell killing and uptake of these targeted nanoparticles.
Materials and Methods
Materials
1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, 1,2-Distearoyl-sn-glycero-3-phosphoethanolamine (DSPE-PEG) 2000, DSPE-PEG 2000 folate were purchased from Avanti Polar Lipids. Inc. (Alabaster, AL), and curcumin (CUR) was obtained from Sigma Aldrich (St. Louis, MO). DSPE-Fluorescein isothiocyanate (FITC) was purchased from Nanosoft Polymers (Winston-Salem, NC). Acetone was ordered from Acros Organics (Waltham, MA). Chloroform was purchased from Sigma Aldrich (St. Louis, MO). All chemicals obtained were analytical grade and were used without any further purification.
Methods
Liposome Synthesis
The components used to make passive and active liposomes (DSPC, cholesterol, DSPE-PEG 2000 (passive) or DSPE-PEG 2000-folate (active). The ratio of lipids used was 2:1:0.2, DSPC:Cholesterol:DSPE-PEG or DSPE-PEG-folate, respectively. The lipid concentration used 10 mg/mL, or 50 mg for a 5-mL organic solution in chloroform, thus the weight of lipid was 50 mg. CUR was approximately 40% by weight of the lipid mass, or 20 mg. The lipids, curcumin and chloroform solvent were combined under the fume hood in a glass vial. The organic solvent chloroform was evaporated using the BUCHI Rotavapor R-3® apparatus under vacuum pressure at a slow rotation rate for 1 hour. The lipid film was rehydrated with deionized water and placed in a 60°C water bath for 24 hours. A LIPEX™ extruder at 60°C under 300 psi compressed nitrogen gas was used to extrude unilamellar vesicles from the multilamellar solution through a 100-nm polycarbonate membrane. Gas extrusion was repeated 11 times for each sample. Overnight dialysis of the liposomal solution was then done at room temperature using MWCO 20 kD dialysis tubing in a 1 L deionized water bath under stirring conditions at 200 rpm. The liposomes were filtered using a 0.22 µm filter and characterized the following day. Empty FITC labeled active and passive liposomes were prepared using the above method, however CUR was not loaded into these liposomes and DSPE-FITC was 0.2% of DSPC by weight.
Characterization
Size, PDI, and zeta potential
The hydrodynamic diameter, polydispersity index (PDI), and zeta potential were measured using a dynamic light scattering (DLS) machine (Malvern Zetasizer Nano - ZS90). For DLS, 60 µL of the sample was diluted with 940 µL of deionized water and the results were measured in a plastic cuvette at 25°C. Samples were tested in triplicate for reproducibility.
Drug encapsulation efficiency
The ThermoScientific NanoDrop 2000c machine was used to measure the absorbance of samples at 425 nm, the maximum wavelength for CUR. Samples were prepared by mixing 10 µL of the liposome sample with 490 µL of 1% Triton-X in deionized water and 500 µL of ethanol to make a 1:1 ethanol:1% Triton X mixture. The drug loading efficiency of the liposomes was calculated using the encapsulation efficiency equation below, whereby the initial synthesis weight of curcumin loaded into liposomes was 20 mg, or 40% of the lipid weight (as previously mentioned in liposome preparation).
Samples were tested in triplicate for reproducibility and the lipid ratio was kept constant for all liposome preparations.
Transmission electron microscopy (TEM)
Samples were prepared for TEM microscopy using the phosphotungstic acid staining technique. An aliquot of liposome sample was placed on a 400 mesh, carbon coated copper grid, allowed to dry for 15 minutes, excess sample removed, followed by another 30-minute drying period, and, finally, addition of 5 uL of 2% phosphotungstic acid (or PTA) solution to negative stain the liposome sample. TEM was performed using a Phillip's EM400T Electron Microscope.
Stability studies
Active and passive liposomes were tested for stability based on size and zeta potential stored at 37℃ in a 1:1 ratio in fetal bovine serum (FBS) over a 3-day period. The temperature and storage media was chosen to simulate human body conditions. A 20 μL aliquot of the sample was added to 980 μL DI water in a plastic cuvette and the sample was placed in the DLS machine to assess size, PDI and zeta potential. Samples were tested in triplicate for reproducibility.
Drug release kinetics
Drug release profiles of the passive and active liposomes were obtained by placing 1 mL samples in 20 kD MWCO dialysis tubing (Spectrum Labs, Rancho Dominguez, CA) in a 1L 1X PBS solution at 37℃ and pH of 7.4 or 5.5. A pH of 5.5 was adjusted by adding 1N HCl to the 1L of 1X PBS solution. The absorbance of curcumin at 425 nm in each sample was measured at selected time points t=0 minutes, t=90 minutes and every 24 hours thereafter until greater than 80% drug release occurred. The samples were prepared by removing the entire contents of the dialysis tubing, measuring the volume, replacing lost volume with DI water to bring the volume to 1 mL, and removing a 20 μL aliquot. The aliquot was diluted with a 1:1 1% Triton-X:ethanol mixture, gently vortexed to rupture the liposome sample, and the absorbance measured using UV-Vis Spectroscopy at 425 nm. Samples were tested in triplicate for reproducibility.
In vitro Studies
Monolayer Cell Culture
MDA-MB-231 and MCF-12A cell lines were obtained from and cultured using the standard protocol provided by American Type Culture Collection (ATCC). The MCF-12A cell line is an adherent, non-tumorigenic, epithelial cell line derived from the mammary glands. MDA-MB-231 cells were cultured in DMEM: Ham's F12 1:1 media with 10% fetal bovine serum and 1% penicillin/streptomycin. MCF-12A cells were grown in DMEM: HAM's F12, 5% horse serum, 20ng/mL EGF, 0.01 mg/mL bovine insulin, 100 ng/mL cholera toxin, 500 ng/mL hydrocortisone and 1% penicillin/streptomycin.
Imaging Experiment
Fluorescence microscopy was utilized to assess the accumulation of CUR in cells using the green fluorescence protein filter setting at fixed transmittance and GFP light intensities which was constant for all imaged cells. GFP has a maximal excitation wavelength at 400 nm, followed by a smaller peak at 475 nm, both of which surround the maximal excitation wavelength for CUR at 425 nm. The imaging was conducted using a monolayer cell culture of TNBC MDA-MB-231 and normal breast epithelial cells MCF-12A. MDA-MB-231 and MCF-12A cells were sub cultured in 35 mm single chambered dishes at a seeding density of 1X106 cells and incubated for 24 hours at 37°C, 5% CO2. Both cell types were dosed with free CUR and CUR encapsulated passive and active liposomes at a concentration of 50 µM the following day. Post 4-hour incubation at 37°C, 5% CO2, the dishes were washed twice with 1X PBS to remove excess drug and imaged in media at 40x magnification to compare CUR uptake in MDA-MB-231 and MCF-12A cells.
Cytotoxicity Studies
MCF-12A and MDA-MB-231 cells were sub cultured in 96 well plates at a seeding density of 40,000 cells/well and incubated for 24 hours at 37ºC, 5% CO2. Cells were dosed with free CUR, passive and active liposomes at concentrations of 5, 10, 25 and 50 µM in their respective media and incubated for 72 hours at 37ºC, 5% CO2. Cytotoxicity studies were performed using the CellTiter 96® AQueous Non-Radioactive Cell Proliferation MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) Assay. MTS cell viability was measured at 490 nm using the Molecular Devices SpectraMax M2 plate reader.
Cell Cycle Analysis
The effects of CUR on cell cycle were investigated using propidium iodide staining in both cell lines previously dosed with the LD50 concentration of CUR for MDA-MB-231 cells obtained through cytotoxicity studies. MDA-MB-231 and MCF-12A cells were sub cultured in 35 mm single chambered dishes at a seeding density of 1X106 cells and incubated for 24 hours at 37°C, 5% CO2. Both cell types were dosed with free CUR and CUR encapsulated passive and active liposomes at a concentration of 19 µM the following day. Post 72-hour incubation at 37°C, 5% CO2, the cells were trypsinized, centrifuged at 500g for 5 minutes, washed with 1x PBS, centrifuged again in 1x PBS at 500 g for 5 minutes, and fixed using 70% ethanol. Propidium iodide (20x concentrated), RNase (200x concentrated) and 1x PBS were added 15 minutes prior to flow cytometry analysis using the procedure from the Abcam Propidium Iodide Flow Cytometry Kit®. Flow cytometry was performed by collecting 10,000 events using the Amnis® FlowSight® Imaging Flow Cytometer using the 488nm excitation laser. Results were analyzed using IDEAS® software.
FITC Uptake studies using Flow Cytometry
Flow cytometry was used to assess the uptake of FITC labeled empty passive and active liposomes in both cell lines. In order to reduce the likelihood of folate interference in cellular uptake of liposomes, 48 hours prior to flow cytometry studies, the cell culture media was switched to folate free media. Folate free media for the MDA-MB-231 cells was comprised of folate free RPMI, 2% FBS, 1% penicillin/streptomycin. Folate free media for the MCF-12A cells was comprised of folate free RPMI, 2% horse serum, 20 ng/mL EGF, 0.01 mg /mL bovine insulin, 100 ng/mL cholera toxin, 500 ng/mL hydrocortisone and 1% penicillin/ streptomycin. Preparation for flow cytometry analysis included addition of TrypLE express to detach cells, followed by centrifugation of the cell suspension at 200x g for 10 minutes. Cell pellets were reconstituted in folate free media, dosed with a fixed quantity of FITC labeled empty liposomes, and incubated at room temperature for 90 minutes on a cell rocker apparatus (to avoid cell settlement). Thereafter, cells were centrifuged again at 200x g for 10 minutes. Removal of the supernatant and addition of 1X PBS followed to wash cells of excess unbound liposomes. The cells were washed 3 times. Formaldehyde was used to fix the cells for 2 hours prior to flow cytometry analysis. Flow cytometry was performed by collecting 10,000 events using the Amnis® FlowSight® Imaging Flow Cytometer using the 488nm excitation laser. Results were analyzed using IDEAS® software.
Statistical Analysis
The results are presented as the average mean ± standard deviation for 3 independent repeats of experiments with triplicate samples in each group, unless stated otherwise. The statistical significance was evaluated using unpaired Student's two-tailed t-test. P<0.05 was considered statistically significant. P<0.05 was represented as *, P<0.01 as ** and P<0.001 as ***.
Results
In this study, we synthesized curcumin-loaded passive liposomes (PL) and actively targeted liposomes (AL). An active targeting moiety was achieved by using folate-functionalized PEG lipid. The folate molecule on the liposomes binds to the folate receptor overexpressed on TNBC cells for targeted drug delivery. The purpose of this study was to compare the efficiency of the liposomal nanoconstructs between non-tumorigenic mammary epithelial cells, MCF-12A, and TNBC cells, MDA-MB-231 [69].
Physicochemical Characterization of Liposomes for Size, PDI, and Zeta Potential
The liposomal constructs synthesized were monodisperse and reproducible. Table 1 below shows that the size of curcumin loaded passive and active liposomes was 120 nm and 130 nm, respectively, with a polydispersity index (PDI) close to 0.1, indicative of a narrow size distribution. The sizes were also analyzed using transmission electron microscopy (TEM). The TEM images verified the DLS results and confirmed the spherical morphology of the two liposomal nanoconstructs as shown in Figure 1 (B) and (C). DLS was also used to measure the zeta potential of PL and AL. Both nanoconstructs showed negative zeta potential of -25 mV and -23 mV for PL and AL, respectively, as Table 1 below and further in Figure 2 (A) (bottom left chart) for zeta potential stability.
Characterization Results for Liposome Size, Polydispersity and Zeta Potential (n=10).
| Size (dnm) | Polydispersity | Zeta Potential (mV) | Encapsulation Efficiency of CUR (%) | |
|---|---|---|---|---|
| PL | 120.16 ± 12 | 0.13 ± 0.06 | -25.10 ± 5.78 | 4.16 ± 0.88% |
| AL | 130.44 ± 16 | 0.14 ± 0.05 | -22.87 ± 6.87 | 4.15 ± 0.98% |
Encapsulation Efficiency
The average encapsulation efficiency that was estimated by UV-Vis spectrometry were 4.16 ± 0.88% and 4.15 ± 0.98% for passive and active curcumin liposomes, respectively, shown above in Table 1. The lipid ratio was kept constant throughout the study where DSPC:Cholesterol:DSPE-PEG or DSPE-PEG-Folate was 2:1:0.1. The encapsulation efficiency was used to assess the improvement of solubility in the liposomal form of CUR. The solubility of CUR in the liposomes was approximately 266 ug/mL, based on the initial weight of CUR loaded in the liposomes (20 mg CUR per 50 mg total lipid content). The average volume of liposomal suspension per batch was approximately 3 mL, yielding an average of 0.266 mg/mL concentration of CUR for both passive and active formulations.
(A) Stability studies are shown for size (top) and zeta potential (bottom) for 3 days (left). Passive (blue bars) had overall smaller sizes than active (green bars) liposomes, while both formulations had similar zeta potential values. (B) Release study results for active and passive CUR loaded liposomes are shown at 37°C (B) pH 5.5 (top-right) and (C) pH 7.4 (bottom-right). Increasing pH appeared to decrease the rate of drug release in both active and passive liposomes significantly. Active liposomes displayed faster initial dissolution at pH 5.5 and 7.4 compared to passive liposomes. These data are mean values ± standard deviation for 3 independent repeats.
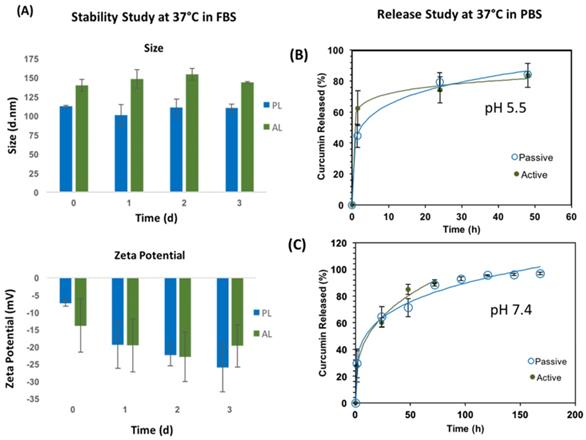
Stability Studies
Stability studies for both liposomal nanoconstructs were performed at 37°C to mimic in vivo testing conditions, as shown in Figure 2 proceeding. Passive and active liposomes were stable for the duration of the 3-day study. Passive liposomes showed relatively smaller sizes than active liposomes as observed in Figure 2 (A) top, although these differences can be attributed to the effects of swelling caused by folate on the surface of active liposomes. Both nanoconstructs demonstrated desirable stability in serum. There was no appreciable change in zeta potential over the duration of the study as well for passive and active alike. It was noted, however, that upon initial addition of serum to the liposomal suspension, the zeta potential appeared to become less negative. The zeta potential thereafter remained more negative compared to day 0, a desirable characteristic for the liposomal preparations for in vivo uses to avoid recognition by the immune system and extend circulation time in the body. Statistical significance testing was performed using the Student's t-test which revealed no statistical difference in stability data or zeta potential for each treatment arm tested. The zeta potential values remained negative for both passive and active, as shown in Figure 2 (A) bottom. A stable size close to 100 nm, PDI values between 0.1 and 0.2, and negative zeta potential are preferable to promote enhanced retention and permeability of nanoparticles by evading recognition by macrophages, reduce protein adsorption on the surface of the nanoparticle which elicit an immune response, and any subsequent immune-mediated deleterious physical side effects in patients.
Drug Release Kinetics
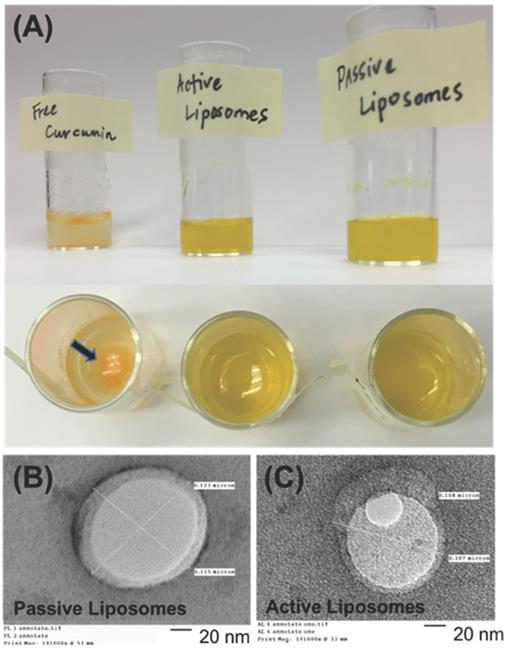
The drug release kinetics were conducted 37°C in 1X PBS at pH 5.5 and 7.4, as shown in Figure 2 (B) and (C), respectively. Excel DD Solver was utilized to find the best fit for the release profiles using various drug delivery rate equations [70]. At 37°C the release profiles for PL and AL at both pH values of 5.5 and 7.4 best fit the Korsmeyer-Peppas (KP) model with an R2 value greater than 0.99 for all data curves. The KP model describes drug dissolution using the power law mathematical model in a simple manner, whereby drug release occurs in a one-dimensional mechanism. Value “n” is the release exponent and “kKP” is the release constant, as indicated in the mathematical equation for % drug release using the KP model that follows.
Values of n 0.5 or less indicate Fickian diffusion, or the flux of mass due to diffusion from an area of higher concentration to an area of lower concentration [71, 72]. The values for the constants and accompanying are shown in Table 2 below.
Constants for drug release kinetics using Korsmeyer-Peppas model (n=3).
| pH = 5.5 kKP | pH = 5.5 n | pH = 7.4 kKP | pH = 7.4 n | |
|---|---|---|---|---|
| PL | 42.774 | 0.186 | 29.681 | 0.247 |
| AL | 59.971 | 0.081 | 23.317 | 0.331 |
Table 3 below shows a comparison between PL and AL CUR release based on pH. The time points used to demonstrate the clear differences in release rate were 50% and 75%. A faster release rate was seen at reduced pH for both passive and active liposomes when compared to a higher pH. At both pH values tested, however, passive liposomes displayed slower release profiles compared to active. The time to 75% drug release was more than 2-fold faster at a lower pH for both PL and AL, while 50% drug release occurred in 5-fold less time than that of the higher pH for PL and approximately 14-fold less time for AL.
Effect of pH on CUR release from liposomal constructs.
| PL pH = 5.5 | PL pH = 7.4 | AL pH = 5.5 | AL pH = 7.4 | |
|---|---|---|---|---|
| Time for 50% CUR release (hours) | 2 | 10 | 0.75 | 11 |
| Time for 75% CUR release (hours) | 22 | 50 | 17 | 39 |
In Vitro Studies
After establishing the reproducible synthesis of the two liposomal constructs, we conducted a series of experiments comparing CUR delivery efficiency of PL and AL in MCF-12A and MDA-MB-231.
Imaging cellular delivery using fluorescence microscopy
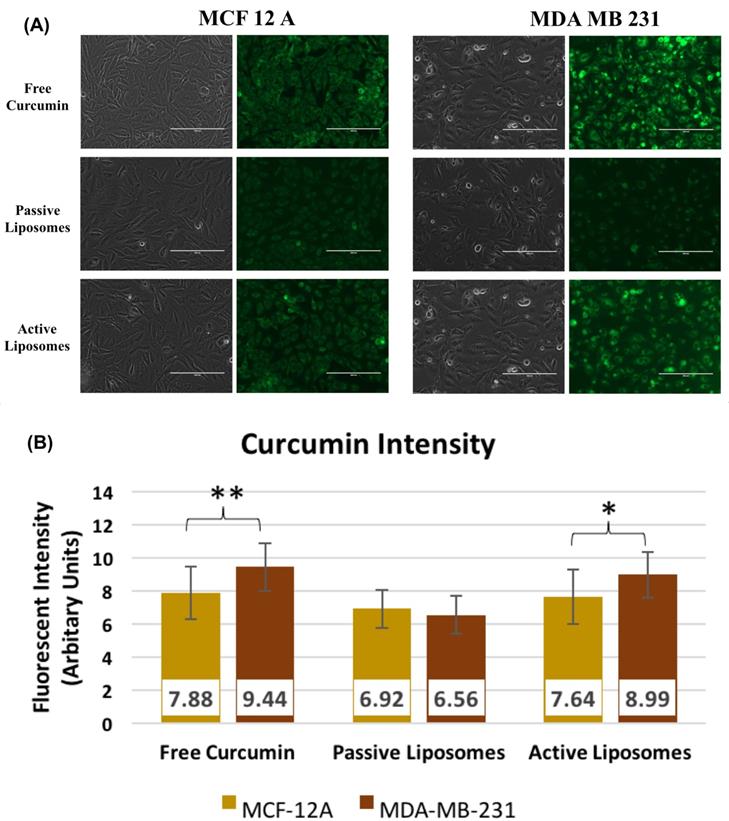
Figure 3 (A) shows the fluorescent intensity variation for comparing CUR uptake in cell lines treated with 50 uM CUR in free and encapsulated form. An incubation period of 90 minutes was used for the study. Figure 3 (A) shows higher CUR fluorescence in MDA-MB-231 (right) compared to MCF-12A (left) indicating that more of CUR was taken up by the TNBC cells compared to normal breast cells. Across the treatment arms, MCF-12A had greater uptake for free CUR followed by AL, while PL showed minimal signal. MDA-MB-231 displayed similar trend as MCF-12A however the CUR signal was brighter. PL again had weak signal, while free CUR and AL had stronger fluorescence. The brighter fluorescence of free CUR compared to other treatment arms may be attributed to the behavior of the free drug in media, whereby free CUR would precipitate out of solution, given CUR insolubility in water, and settle at the bottom of the dish where the cells are adhered. Passive diffusion of free CUR would then result in an enhanced fluorescence. The fluorescent intensity values shown in Figure 3 (B) do not display a remarkable difference across treatment arms, however free CUR and AL appeared to have slightly higher values in MDA-MB-231 cells. MCF-12A cells had similar results for intensity across all treatment arms.
(A) Fluorescence microscopy results comparing cell lines and the uptake of 50 uM free drug CUR, PL and AL after 90 minutes incubation with PL or AL. Images were taken at 40x magnification using GFP filter for CUR. The left side images under each cell type are transmittance images, while right side images are GFP filter images showing CUR fluorescence in cells. Images shown are representative of 3 independent repeats. (B) Fluorescent intensity values for CUR corresponding to the images shown in (A) to compare the differences found between cell lines. * p<0.05, **p<0.01, and ***p<0.005 using Student's t-test. These data are mean values ± standard deviation for 3 independent repeats.
Cytotoxicity Results
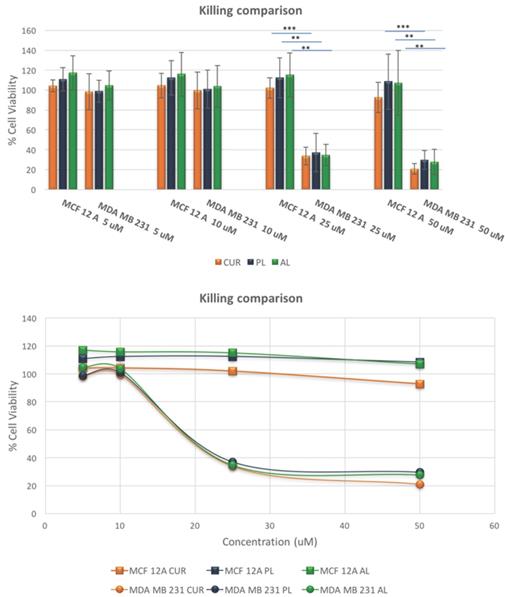
After confirming the difference in the CUR uptake, we investigated the killing efficacy of the two liposomal constructs on MCF-12A and MDA-MA-231 as shown in Figure 4 below. The cytotoxicity experiments were performed over a period of 72-hours for each treatment arm, free CUR, PL and AL at concentrations of 5 uM, 10 uM, 25 uM and 50 uM. MCF-12A showed no cell death compared to no treatment, while MDA-MB-231 demonstrated LD50 around 19 uM for all treatment arms. At lower concentrations, 5uM and 10 uM, both cell lines showed no cell death for free CUR, PL and AL. Cytotoxicity was mainly observed at 25 uM and 50 uM for MDA-MB-231, unlike MCF-12A which showed cell viability closer to 100%. The difference in the killing efficiency for both cell lines was statistically significant for all treatments.
Flow cytometry experiments
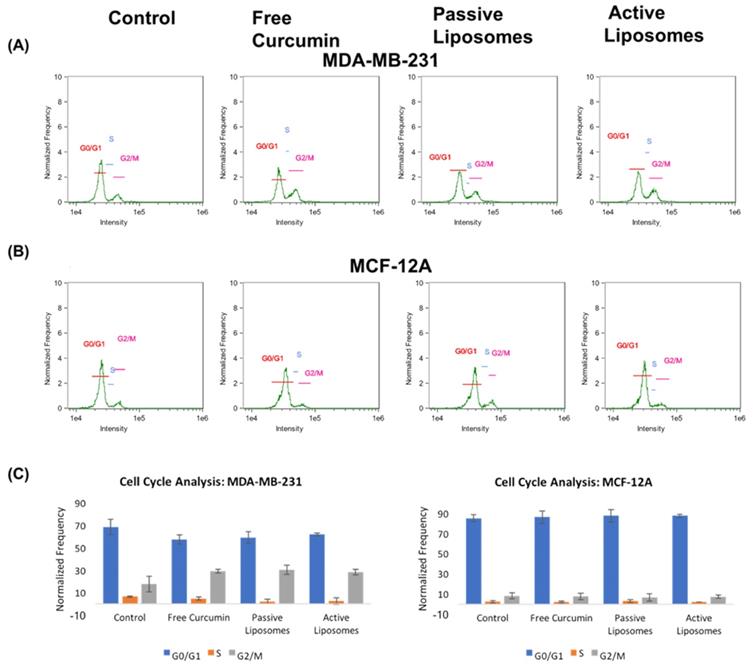
While the imaging and killing studies showed the specificity of CUR towards cancer cells versus non-malignant cells, a quantitative analysis was needed to assess the effects of CUR on cell cycle, as well as the targeting behavior of the liposomal constructs. The cell cycle analysis results are shown in Figure 5. While MCF-12A (Figure 5 (B)) cells appeared to have no notable difference in the percentages of cells in G0/G1, S and G2/M across all treatment arms when compared to the no treatment group, the MDA-MB-231 (Figure 5 (A)) cells showed an increase in the number of cells arrested in the G2/M phase, compared to the no treatment group. CUR did not appear to alter the cell cycle of normal cells, while cancer cells demonstrated a shift in cell cycle results upon treatment with CUR, both in free and encapsulated formulation. There was no apparent difference, however, in the effect of free CUR, PL, and AL on MDA-MB-231 when compared to one another (further shown in Figure 5 (C) bar graph representation), the values to which in shown in the supplementary table S1.
Killing comparison results show the differences in cytotoxic effects of free drug CUR, PL and AL in normal breast epithelial cells and TNBC cells at various concentrations. A significant change in cytotoxicity between cell lines, where TNBC cells had dramatically less cell viability among all treatment arms compared to 12A normal cells. Statistical significance was found at 25 uM and 50 uM concentrations. * p<0.05, **p<0.01, and ***p<0.005 using Student's t-test. These data are mean values ± standard deviation for 3 independent repeats.
Cell cycle analysis using propidium iodide revealed cellular arrest in G2/M phase in CUR treated groups (A) for MDA-MB-231 cells, while (B) MCF-12A cells did not display appreciable difference in the frequency of cells in G2/M after incubation with CUR treatment arms. The overall results in (C) demonstrate these differences between cell lines comparing the frequency of cells found in each phase of the cell cycle as a percent of total cells gated using flow cytometry.
Flow cytometry regions shown in fluorescence intensity results and their corresponding cell population with specific FITC intensity.
| Region | Intensity of FITC |
|---|---|
| 1 | 0-5,000 |
| 2 | 5,000-10,000 |
| 3 | 10,000-30,000 |
| 4 | 30,000-130,000 |
| 5 | 130,000-1,000,000 |
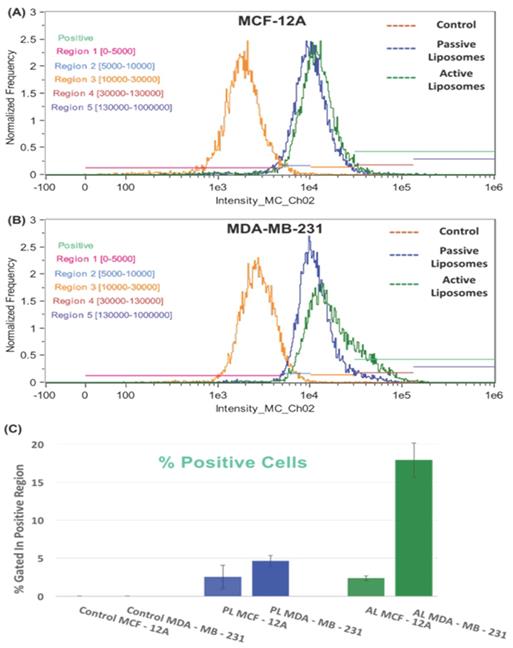
To study the targeting potential of each liposomal formulation, we decided to conduct a flow cytometry experiment (Figure 6). The treatment arms in Figures 6 (A) and 6 (B) are indicated by no treatment (orange peak), passive liposomes (blue peak) and active liposomes (green peak). The flow cytometry results showed increased uptake of active empty FITC labeled liposomes in MDA-MB-231 cells compared to MCF-12A, whereas no difference was observed for passive liposomes in both the cell lines.
Furthermore, the FITC intensity (x-axis) was divided into 5 regions as described in Table 4 above, where the “positive uptake” cell population (light green) was defined as cells in regions 4 and 5, shown by maroon and purple horizontal lines in Figures 6 (A) and 6 (B), shown to the right. Figure 6 (C) displays the differences in % positive cells based on the uptake of liposomes. The relative % positive cells were similar between cell lines for passive liposomes, however active liposomes showed approximately 15% difference in % positive cells. Within the MDA-MB-231 cell line, there was a marked difference between empty PL and empty AL showing 4% and 18% positive cell uptake, respectively, the data to which is shown in the supplementary table S.2. This proves that the mechanism of uptake is different between PL and AL whereby receptor mediated endocytosis resulted in increased % positive cells when treated with empty FITC AL.
The fluorescence intensity of FITC is shown for the treatment arms (no treatment (orange), passive empty liposomes (blue), and active empty liposomes (green)) for (A) MCF-12A cells and (B) MDA-MB-231 cells. The percent positive cells for FITC fluorescence are also shown in (C) as a histogram to demonstrate the differences between cell lines for cellular uptake of passive and active empty liposomes. A shift can be seen in the fluorescence intensity in (B) for MDA-MB-231 uptake of active empty liposomes where increased uptake occurred, compared to normal cells in (A). These data are mean values ± standard deviation for 2 independent repeats.
Discussion
Curcumin is a natural polyphenol compound that has shown anti-inflammatory and anti-carcinogenic abilities [20, 30]. As of 2017 there are 56 clinical trials reported on clinicialtrials.gov pertaining to using curcumin as a monotherapy or in combination for cancer treatment. This data exhibits the potential of curcumin related therapy to get clinical approval for cancer treatment. While its ability as a chemotherapeutic and chemopreventative drug is indisputable, its hydrophobicity renders curcumin ineffective due to poor systemic bioavailability and rapid clearance from the body [30, 37]. The purpose of this study was to address these issues by encapsulating curcumin in folate targeted liposomes and studying the difference in its therapeutic efficiency between nonmalignant cells and cancer cells. Our hypothesis was that folate targeted liposomes only aid curcumin's specificity to fight against cancer. To test this hypothesis, in vitro cellular uptake and killing were designed to test the therapeutic specificity of curcumin loaded folate targeted liposomes. The representative cell lines used in this study were MCF-12A, as non-malignant mammary epithelial cell line and MDA-MB-231, as triple negative breast cancer cell line. While several studies have demonstrated therapeutic efficacy of curcumin loaded folate targeted liposomes either as a monotherapy or combination therapy [73, 74], we wanted to design an experiment to conduct a direct comparison of liposomal CUR between nonmalignant cells and cancer cells.
Two types of liposomal nanoconstructs were engineered for the purpose of this study, folate targeted curcumin loaded liposomes (AL) and non-targeted curcumin loaded liposomes (PL) (as a control). The liposomes were synthesized using a mature process of thin-film hydration which produced reproducible, consistent and clinically preferable unilamellar, monodisperse liposomal constructs, based on the average size of 125 nm and PDI close to 0.1 for passive and active formulations [75]. Monodispersity in nanoparticles is necessary to avoid causing an immune response [76-78]. The negative zeta potential of both PL and AL indicates that they have better systemic stability, and are less likely to cause immunogenicity in vivo [74, 79, 80]. Nanoparticles which are close to 100 nm can cross the blood-brain barrier and evade the reticuloendothelial system, resulting in prolonged circulation time, thus making smaller liposomes even more attractive as a biocompatible drug delivery system [5, 81]. The morphological attributes of the liposomes were confirmed using transmission electron microscopy, TEM, which validated the DLS findings for liposome size and showed that they were spherical in shape (as shown previously in Figure 1 (B) images).
The release studies were performed at 37ºC (pH 7.4 and 5.5) in 1X PBS to determine the profile of CUR release from the liposomes, whereby 37℃ was chosen to simulate physiological conditions. The active liposomes demonstrated faster drug release kinetics than passive liposomes at 37ºC at later time points. In vivo testing of liposomal formulations would be expected to follow the release kinetics found in the 37ºC testing parameter, although interactions with serum proteins may increase drug release rates due to protein interactions and subsequent disturbances in liposome stability over time in the bloodstream. The liposomes prepared in this study, however, do demonstrate acceptable stability and sustained release over several days, a key attribute of the liposomal design in drug delivery applications in vivo.
The effect on release of CUR at reduced pH is relevant to the behavior of the drug in relation to the acidic endosomal environment. CUR rapidly degrades as pH increases in free drug form, as shown in previous work by Feng et al., therefore liposomal encapsulation is ideal to reduce the effects of pH in drug delivery [5]. An increase in drug release at lower pH was observed in our study, among other investigations as well. This increase in drug release is due to the collapse of the transmembrane pH gradient of the liposomes. After being internalized by endosomes, the liposomes will release CUR. Lee et al also observed an increase in the release of CUR at lower pH using folate-tagged liposomes [82, 83]. The folate decorated liposomes used in the study may have had faster release because folate incorporation in the lipid bilayer surface results in reduced rigidity and stability of the lipid membrane [82]. Folate lipids also possess more heterogeneity and subsequent poor lipid packing, thus causing higher permeability and susceptibility to the effects of reduced pH [82]. The general release profile of active CUR liposomes displayed faster release at higher temperature and reduced pH and could be attributed to the characteristic traits of the lipid bilayer aforementioned. Stability and drug release studies were performed to confirm that the liposomal nanoconstructs were durable and provide gradual drug escape, thus ensuring not only the shelf life of the product, but also gradual liposome degradation and desirable release profiles. Rapid degradation in the blood stream would be undesirable due to increased toxicity and less cellular uptake in tumor tissue, exacerbated by the mononuclear phagocytic system (MPS) which is a primary clearance route for intravenous liposomal drug delivery, thus making release kinetics critical testing parameters useful for clinical application [84].
The size, PDI, and zeta potential results for stability showed that both passive and active liposomes are stable at body temperature conditions of 37°C in FBS. Ligand modification of the liposomes did not appear to impede the stability of liposomes in this study. The effects of protein adsorption and charge repulsion on zeta potential measurements are evident, as the zeta potential fluctuated from less negative to increasing negativity as the stability study in FBS progressed for several days. The stability of liposomes in suspensions is dependent on the repulsive interactions which must be similar to the magnitude and range of the van der Waals force, or the weak, induced electrical intermolecular forces between atoms [85, 86]. Electrostatic and steric stabilization prevent the close interaction of liposomes, while aggregation can occur when proteins are adsorbed on the surface and alter the charge. The variation of charge indicated by our results may be a result of these interactions with proteins in serum and subsequent decrease in negativity upon initial exposure to FBS on day 1, while thereafter repulsion and steric hindrance among liposomes in suspension overcame such interactions and drove zeta potentials back to strong negative values, indicating stability over time [85].
After ensuring the physicochemical characteristics, the liposomes were tested in a series of in vitro cell culture experiments. We investigated the ability of PL and AL to deliver CUR to both cell lines using fluorescence microscopy. A 90-minute incubation period was set for the imaging study because the release rate at 37ºC of both PL and AL was the same at this time point (~28%). This was critical to minimize the effect of differences in curcumin release from the two liposomal constructs. CUR fluorescence was greater in MDA-MB-231 for all treatment arms compared to MCF-12A primarily because of difference in the metabolism such as cell doubling period, morphology and rate of endocytosis [87], however a similar trend was observed for MCF-12A. The brighter fluorescence of free CUR compared to other treatment arms may be attributed to the behavior of the free drug in media, whereby free CUR would precipitate out of solution, given CUR insolubility in aqueous solution, and settle at the bottom of the dish where the cells are adhered. Passive diffusion of free CUR would then result in an enhanced fluorescence. CUR uptake in cells when treated with PL is mainly via diffusion of released drug in the extracellular space, while for AL treated cells, in addition to passive diffusion, an additional mechanism of uptake is via folate receptor-mediated endocytosis (RME). All cells have folate receptors, which enables folate based targeting. However, folate receptors are overexpressed in several tumor cells including MDA-MB-231[88], which is another reason why AL treated MDA-MB-231 cells have stronger signal compared to AL treated MCF-12A.
Next, we tested the efficacy of the liposomal constructs to not induce cytotoxicity in both the cell lines, but also see how CUR affects the cell cycle of normal and cancerous cells. The killing studies were conducted over a period of 72 hours at 4 different concentrations ranging from 5 uM to 50 uM. The LD50 values were not achieved for MCF-12A, while MDA-MB-231 showed LD50 around 19 uM for all three treatment arms. Moreover, at 50 uM the cell viability for MCF-12A remained similar to no treatment unlike MDA-MB-231, indicating that curcumin loaded liposomes at such concentrations can be more cytotoxic to cancer cells compared to normal cells. Moreover, the AL did not alter the therapeutic potential and selectively of CUR, but only helped in improving the bioavailability of the drug to enhance CUR potential as an anticancer drug. While the results looked promising with the in vitro model, it will be interesting to achieve similar outcome with 3D models using similar and different cancer types to closely simulate in vivo tumor penetration of curcumin-encapsulated liposomes [89, 90]. As a future perspective for this research 3D cell culture experiments would be useful to validate the findings of the 2D monolayer work presented here. In vivo studies will also provide crucial evidence of the ability to actively target tumor cells, however the in vivo study is outside the scope of the research presented. We sought to investigate the foundation framework of the comparison effects of free versus encapsulated CUR formulations between cell lines. In vivo studies on the effects of curcumin encapsulation have been performed previously in other cancer types [23, 27, 91-93]. Dhule et al reported using mesenchymal stem cells that non-malignant cells are affected by CUR such that arrest in G0 occurs reversibly without apoptosis occurring, while also emphasizing how cancerous cell growth rate is critical in the uptake of liposomes and subsequent drug cytotoxicity. Multi drug resistance in cancer cells from mutations also contributes to insensitivity of tumor cells to pharmaceutical intervention. Varied growth rates between normal and cancerous cells affects drug sensitivity as well, whereby a cancer cell has more opportunity to evade destruction over time through ongoing genetic disturbances.
The effects of CUR on cell cycle may be attributed to what has been studied previously in TNBC using polymeric based CUR encapsulation [94]. CUR has been found to cause cell cycle arrest at G1/S and G2/M phase in TNBC cells. Cell cycle arrest has been associated with the production of reactive oxygen species resulting from CUR treatment leads to DNA damage and reductions in CDK2, CDK4, cyclin D1 and cyclin E because of p38-MAPK activation [94]. Cell cycle arrest in the G2/M phase has been noted in previous work by Dhule et al which compared the use of curcumin liposomes, coencapsulated curcumin and C6 ceramide liposomes, in addition to folate decorated CUR-C6 ceramide combination liposomes in the treatment of osteosarcoma in vitro and in vivo. Dhule et al noted that the effect of curcumin-loaded liposomes exclusively induced the G2/M arrest by upregulating cyclin B1 specifically [27]. Ucisik, M.H. et. al. also showed that encapsulated curcumin increases cell cycle arrest at G2/M phases comparable to the results we found with MDA-MB-231 cells [43].
While we successfully determined that liposomal formulations were just as effective as free CUR in killing cancer cells, the 72-hour killing study did not show an appreciable difference in cytotoxicity between PL and AL. Although monolayer cell culture technique offers valuable insight on the drug efficacy, it comes with several limitations in understanding the interactions between the nanoparticles, the released drug(s) and the adherent cells, which makes it difficult to understand cellular uptake mechanisms [95]. This is mainly because, the nanoparticles and the released drug can bind to or diffuse through the cells respectively, without any restrictions as observed in in vivo models [27, 95, 96]. However, within the monolayer setup, the specificity of folate decorated nanosystems are often confirmed by conducting competition assays with free folic acid. In a recent study published, Jones, A.K. and colleagues showed that multivalency of the targeted nanosystems offers higher binding affinity to the overexpressed FAα than monovalent folic acid. This leads to questionable inference of the results obtained via competition assay rendering inconclusive indication of targeting potential of the folate functionalized nanosystems [97]. Hence, for the purposes of our study, we utilized fluorescent labeled empty liposomes to quantitatively distinguish between the uptake of our folate decorated and non-decorated liposomes. This was done by synthesizing empty (non-CUR loaded) passive and active fluorescein isothiocyanate (FITC) labeled liposomes to test the uptake using flow cytometry. Empty liposomes were used to demonstrate targeted potential of AL to folate receptors and to eliminate any possible difference in the curcumin release rate from PL and AL. Thus, the mere distinguishing factors between FITC labeled empty passive and active liposomes is folate functionalization. The following pivotal information can be inferred from the flow experiments: a) Compared to MCF-12A, MDA-MB-231 showed greater FITC intensity, b) AL treated cells showed higher FITC intensity compared to PL for both the cell lines, indicating the specificity of folate functionalized liposomes to folate receptors, verifying that overexpressed folate receptors on tumor cells (compared to normal cells) make our liposomal construct beneficial for treatment of cancer cells. While these results showed strong evidence for our selected cell line, folate targeted liposomes can aid in curcumin therapy for other cancer types as well.
Liposomes are useful vehicles to deliver CUR because of their ability to improve bioavailability and solubility of curcumin. They can be used as a potential targeted delivery system by folate-ligand surface modification, gradual release of CUR in the body, and subsequent improved efficacy of treatment in cancer patients [5, 39, 51, 98, 99]. It is important to understand the behavior of such surface modified drug delivery systems in both cancer and non-malignant cells as a preemptive means to postulate and further test said therapeutics in vivo. This study showed that folate-targeted liposomes do not alter the selective cytotoxicity behavior of curcumin in our comparative in vitro cell study. In fact, the use of folate in specializing CUR liposomes was verified in our studies to be an advantageous instrument for improving drug delivery and cellular uptake in cancerous cells. In conclusion, the comparison between MDA-MB-231 and MCF-12A confirmed the difference in the efficacy of curcumin for the both the cell lines when subjected to similar conditions. These results highlight the importance of folate-targeted liposomes and its role in delivery of curcumin for cancer treatment.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
The authors acknowledge funding from the National Cancer Institute, a part of the National Institutes of Health (Award # R00CA153948). The authors would also like to acknowledge Dr. Earl Ada and Jack Lepine from the UMass Lowell Biomolecular Characterization Lab for their advice in microscopy and flow cytometry, as well as peers in Dr. Prakash Rai's nanotechnology lab, including Shailee Paliwal, Bobbi Yassini, Leo Kutlowski and Lewis Kerrebrock for their assistance in stability study characterization.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Miller JM, Thompson JK, MacPherson MB, Beuschel SL, Westbom CM, Sayan M. et al. Curcumin: a double hit on malignant mesothelioma. Cancer Prev Res (Phila). 2014;7:330-40
2. Irving GR, Howells LM, Sale S, Kralj-Hans I, Atkin WS, Clark SK. et al. Prolonged biologically active colonic tissue levels of curcumin achieved after oral administration-a clinical pilot study including assessment of patient acceptability. Cancer Prev Res (Phila). 2013;6:119-28
3. Sun D, Zhuang X, Xiang X, Liu Y, Zhang S, Liu C. et al. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther. 2010;18:1606-14
4. Dance-Barnes ST, Kock ND, Moore JE, Lin EY, Mosley LJ, D'Agostino RB Jr. et al. Lung tumor promotion by curcumin. Carcinogenesis. 2009;30:1016-23
5. Feng T, Wei Y, Lee RJ, Zhao L. Liposomal curcumin and its application in cancer. Int J Nanomedicine. 2017;12:6027-44
6. Hussain Z, Thu HE, Amjad MW, Hussain F, Ahmed TA, Khan S. Exploring recent developments to improve antioxidant, anti-inflammatory and antimicrobial efficacy of curcumin: A review of new trends and future perspectives. Mater Sci Eng C Mater Biol Appl. 2017;77:1316-26
7. Morimoto T, Sunagawa Y, Katanasaka Y, Hirano S, Namiki M, Watanabe Y. et al. Drinkable preparation of Theracurmin exhibits high absorption efficiency-a single-dose, double-blind, 4-way crossover study. Biol Pharm Bull. 2013;36:1708-14
8. Olivera A, Moore TW, Hu F, Brown AP, Sun A, Liotta DC. et al. Inhibition of the NF-kappaB signaling pathway by the curcumin analog, 3,5-Bis(2-pyridinylmethylidene)-4-piperidone (EF31): anti-inflammatory and anti-cancer properties. Int Immunopharmacol. 2012;12:368-77
9. Ghosh A, Banerjee T, Bhandary S, Surolia A. Formulation of nanotized curcumin and demonstration of its antimalarial efficacy. Int J Nanomedicine. 2014;9:5373-87
10. Yallapu MM, Nagesh PK, Jaggi M, Chauhan SC. Therapeutic Applications of Curcumin Nanoformulations. AAPS J. 2015;17:1341-56
11. Lee WH, Loo CY, Bebawy M, Luk F, Mason RS, Rohanizadeh R. Curcumin and its derivatives: their application in neuropharmacology and neuroscience in the 21st century. Curr Neuropharmacol. 2013;11:338-78
12. Yang Z, Sun N, Cheng R, Zhao C, Liu Z, Li X. et al. pH multistage responsive micellar system with charge-switch and PEG layer detachment for co-delivery of paclitaxel and curcumin to synergistically eliminate breast cancer stem cells. Biomaterials. 2017;147:53-67
13. Xu Y, Asghar S, Yang L, Chen Z, Li H, Shi W. et al. Nanoparticles based on chitosan hydrochloride/hyaluronic acid/PEG containing curcumin: In vitro evaluation and pharmacokinetics in rats. Int J Biol Macromol. 2017;102:1083-91
14. Cui T, Zhang S, Sun H. Co-delivery of doxorubicin and pH-sensitive curcumin prodrug by transferrin-targeted nanoparticles for breast cancer treatment. Oncol Rep. 2017;37:1253-60
15. Yang Z, Sun N, Cheng R, Zhao C, Liu J, Tian Z. Hybrid nanoparticles coated with hyaluronic acid lipoid for targeted co-delivery of paclitaxel and curcumin to synergistically eliminate breast cancer stem cells. Journal of Materials Chemistry B. 2017;5:6762-75
16. Subramani PA, Panati K, Narala VR. Curcumin Nanotechnologies and Its Anticancer Activity. Nutr Cancer. 2017;69:381-93
17. Kalashnikova I, Mazar J, Neal CJ, Rosado AL, Das S, Westmoreland TJ. et al. Nanoparticle delivery of curcumin induces cellular hypoxia and ROS-mediated apoptosis via modulation of Bcl-2/Bax in human neuroblastoma. Nanoscale. 2017;9:10375-87
18. Kang M, Ho JN, Kook HR, Lee S, Oh JJ, Hong SK. et al. Theracurmin(R) efficiently inhibits the growth of human prostate and bladder cancer cells via induction of apoptotic cell death and cell cycle arrest. Oncol Rep. 2016;35:1463-72
19. Strong R, Miller RA, Astle CM, Baur JA, de Cabo R, Fernandez E. et al. Evaluation of resveratrol, green tea extract, curcumin, oxaloacetic acid, and medium-chain triglyceride oil on life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2013;68:6-16
20. Wang K, Zhang C, Bao J, Jia X, Liang Y, Wang X. et al. Synergistic chemopreventive effects of curcumin and berberine on human breast cancer cells through induction of apoptosis and autophagic cell death. Sci Rep. 2016;6:26064
21. Scarano W, de Souza P, Stenzel MH. Dual-drug delivery of curcumin and platinum drugs in polymeric micelles enhances the synergistic effects: a double act for the treatment of multidrug-resistant cancer. Biomater Sci. 2015;3:163-74
22. Wang H, Ke F, Mararenko A, Wei Z, Banerjee P, Zhou S. Responsive polymer-fluorescent carbon nanoparticle hybrid nanogels for optical temperature sensing, near-infrared light-responsive drug release, and tumor cell imaging. Nanoscale. 2014;6:7443-52
23. Sarisozen C, Abouzeid AH, Torchilin VP. The effect of co-delivery of paclitaxel and curcumin by transferrin-targeted PEG-PE-based mixed micelles on resistant ovarian cancer in 3-D spheroids and in vivo tumors. Eur J Pharm Biopharm. 2014;88:539-50
24. Zhang Y, Yang C, Wang W, Liu J, Liu Q, Huang F. et al. Co-delivery of doxorubicin and curcumin by pH-sensitive prodrug nanoparticle for combination therapy of cancer. Sci Rep. 2016;6:21225
25. Chen D, Dong X, Qi M, Song X, Sun J. Dual pH/redox responsive and CD44 receptor targeting hybrid nano-chrysalis based on new oligosaccharides of hyaluronan conjugates. Carbohydr Polym. 2017;157:1272-80
26. Yang L, Gao S, Asghar S, Liu G, Song J, Wang X. et al. Hyaluronic acid/chitosan nanoparticles for delivery of curcuminoid and its in vitro evaluation in glioma cells. Int J Biol Macromol. 2015;72:1391-401
27. Dhule SS, Penfornis P, He J, Harris MR, Terry T, John V. et al. The combined effect of encapsulating curcumin and C6 ceramide in liposomal nanoparticles against osteosarcoma. Mol Pharm. 2014;11:417-27
28. Sercombe L, Veerati T, Moheimani F, Wu SY, Sood AK, Hua S. Advances and Challenges of Liposome Assisted Drug Delivery. Front Pharmacol. 2015;6:286
29. Monteiro N, Martins A, Reis RL, Neves NM. Liposomes in tissue engineering and regenerative medicine. J R Soc Interface. 2014;11:20140459
30. Zaman MS, Chauhan N, Yallapu MM, Gara RK, Maher DM, Kumari S. et al. Curcumin Nanoformulation for Cervical Cancer Treatment. Sci Rep. 2016;6:20051
31. Banerjee S, Sahoo AK, Chattopadhyay A, Ghosh SS. Recombinant IkappaBalpha-loaded curcumin nanoparticles for improved cancer therapeutics. Nanotechnology. 2014;25:345102
32. Gong G, Pan Q, Wang K, Wu R, Sun Y, Lu Y. Curcumin-incorporated albumin nanoparticles and its tumor image. Nanotechnology. 2015;26:045603
33. Pawar YB, Purohit H, Valicherla GR, Munjal B, Lale SV, Patel SB. et al. Novel lipid based oral formulation of curcumin: development and optimization by design of experiments approach. Int J Pharm. 2012;436:617-23
34. Wahlang B, Kabra D, Pawar YB, Tikoo K, Bansal AK. Contribution of formulation and excipients towards enhanced permeation of curcumin. Arzneimittelforschung. 2012;62:88-93
35. Chen Y, Wu Q, Zhang Z, Yuan L, Liu X, Zhou L. Preparation of curcumin-loaded liposomes and evaluation of their skin permeation and pharmacodynamics. Molecules. 2012;17:5972-87
36. Pandelidou M, Dimas K, Georgopoulos A, Hatziantoniou S, Demetzos C. Preparation and characterization of lyophilised egg PC liposomes incorporating curcumin and evaluation of its activity against colorectal cancer cell lines. J Nanosci Nanotechnol. 2011;11:1259-66
37. Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007;4:807-18
38. He W, Zou C, Tian Z, Tan W, Shen W, Chen J. et al. Nasopharyngeal carcinoma treated with bevacizumab combined with paclitaxel liposome plus cisplatin: a case report and literature review. Onco Targets Ther. 2017;10:67-72
39. Mondal G, Barui S, Saha S, Chaudhuri A. Tumor growth inhibition through targeting liposomally bound curcumin to tumor vasculature. J Control Release. 2013;172:832-40
40. Rudokas M, Najlah M, Alhnan MA, Elhissi A. Liposome Delivery Systems for Inhalation: A Critical Review Highlighting Formulation Issues and Anticancer Applications. Med Princ Pract. 2016;25(Suppl 2):60-72
41. Mahmud M, Piwoni A, Filipczak N, Janicka M, Gubernator J. Long-Circulating Curcumin-Loaded Liposome Formulations with High Incorporation Efficiency, Stability and Anticancer Activity towards Pancreatic Adenocarcinoma Cell Lines In Vitro. PLoS One. 2016;11:e0167787
42. Kitajima H, Komizu Y, Ichihara H, Goto K, Ueoka R. Hybrid liposomes inhibit tumor growth and lung metastasis of murine osteosarcoma cells. Cancer Med. 2013;2:267-76
43. Ucisik MH, Kupcu S, Schuster B, Sleytr UB. Characterization of CurcuEmulsomes: nanoformulation for enhanced solubility and delivery of curcumin. J Nanobiotechnology. 2013;11:37
44. Yallapu MM, Jaggi M, Chauhan SC. Curcumin nanoformulations: a future nanomedicine for cancer. Drug Discov Today. 2012;17:71-80
45. De M, Ghosh S, Sen T, Shadab M, Banerjee I, Basu S. et al. A Novel Therapeutic Strategy for Cancer Using Phosphatidylserine Targeting Stearylamine-Bearing Cationic Liposomes. Mol Ther Nucleic Acids. 2018;10:9-27
46. Affram K, Udofot O, Singh M, Krishnan S, Reams R, Rosenberg J. et al. Smart thermosensitive liposomes for effective solid tumor therapy and in vivo imaging. PLoS One. 2017;12:e0185116
47. Miyazawa T, Kamiyoshihara R, Shimizu N, Harigae T, Otoki Y, Ito J. et al. Amadori-glycated phosphatidylethanolamine enhances the physical stability and selective targeting ability of liposomes. R Soc Open Sci. 2018;5:171249
48. Storka A, Vcelar B, Klickovic U, Gouya G, Weisshaar S, Aschauer S. et al. Safety, tolerability and pharmacokinetics of liposomal curcumin in healthy humans. Int J Clin Pharmacol Ther. 2015;53:54-65
49. Milano F, Mari L, van de Luijtgaarden W, Parikh K, Calpe S, Krishnadath KK. Nano-curcumin inhibits proliferation of esophageal adenocarcinoma cells and enhances the T cell mediated immune response. Front Oncol. 2013;3:137
50. Kanai M, Otsuka Y, Otsuka K, Sato M, Nishimura T, Mori Y. et al. A phase I study investigating the safety and pharmacokinetics of highly bioavailable curcumin (Theracurmin) in cancer patients. Cancer Chemother Pharmacol. 2013;71:1521-30
51. Sneider A, Jadia R, Piel B, VanDyke D, Tsiros C, Rai P. Engineering Remotely Triggered Liposomes to Target Triple Negative Breast Cancer. Oncomedicine. 2017;2:1-13
52. Hou Z, Gattoc L, O'Connor C, Yang S, Wallace-Povirk A, George C. et al. Dual Targeting of Epithelial Ovarian Cancer Via Folate Receptor alpha and the Proton-Coupled Folate Transporter with 6-Substituted Pyrrolo[2,3-d]pyrimidine Antifolates. Mol Cancer Ther. 2017;16:819-30
53. Zhang X, Liu N, Shao Z, Qiu H, Yao H, Ji J. et al. Folate-targeted nanoparticle delivery of androgen receptor shRNA enhances the sensitivity of hormone-independent prostate cancer to radiotherapy. Nanomedicine. 2017;13:1309-21
54. Rosiere R, Gelbcke M, Mathieu V, Van Antwerpen P, Amighi K, Wauthoz N. New dry powders for inhalation containing temozolomide-based nanomicelles for improved lung cancer therapy. Int J Oncol. 2015;47:1131-42
55. Yin C, Hong B, Gong Z, Zhao H, Hu W, Lu X. et al. Fluorescent oligo(p-phenyleneethynylene) contained amphiphiles-encapsulated magnetic nanoparticles for targeted magnetic resonance and two-photon optical imaging in vitro and in vivo. Nanoscale. 2015;7:8907-19
56. Ndong C, Toraya-Brown S, Kekalo K, Baker I, Gerngross TU, Fiering SN. et al. Antibody-mediated targeting of iron oxide nanoparticles to the folate receptor alpha increases tumor cell association in vitro and in vivo. Int J Nanomedicine. 2015;10:2595-617
57. Ab O, Whiteman KR, Bartle LM, Sun X, Singh R, Tavares D. et al. IMGN853, a Folate Receptor-alpha (FRalpha)-Targeting Antibody-Drug Conjugate, Exhibits Potent Targeted Antitumor Activity against FRalpha-Expressing Tumors. Mol Cancer Ther. 2015;14:1605-13
58. Chang LC, Hsieh MT, Yang JS, Lu CC, Tsai FJ, Tsao JW. et al. Effect of bis(hydroxymethyl) alkanoate curcuminoid derivative MTH-3 on cell cycle arrest, apoptotic and autophagic pathway in triple-negative breast adenocarcinoma MDA-MB-231 cells: An in vitro study. Int J Oncol. 2018;52:67-76
59. Liu W, Fu X, Yang Z, Li S, Cao Y, Li Q. et al. Moderate intermittent negative pressure increases invasiveness of MDA-MB-231 triple negative breast cancer cells. Breast. 2017;38:14-21
60. Milczarek M, Wiktorska K, Mielczarek L, Koronkiewicz M, Dabrowska A, Lubelska K. et al. Autophagic cell death and premature senescence: New mechanism of 5-fluorouracil and sulforaphane synergistic anticancer effect in MDA-MB-231 triple negative breast cancer cell line. Food Chem Toxicol. 2017;111:1-8
61. Etti IC, Abdullah R, Kadir A, Hashim NM, Yeap SK, Imam MU. et al. The molecular mechanism of the anticancer effect of Artonin E in MDA-MB 231 triple negative breast cancer cells. PLoS One. 2017;12:e0182357
62. Huang L, Li A, Liao G, Yang F, Yang J, Chen X. et al. Curcumol triggers apoptosis of p53 mutant triple-negative human breast cancer MDA-MB 231 cells via activation of p73 and PUMA. Oncol Lett. 2017;14:1080-8
63. Wilder PT, Weber DJ, Winstead A, Parnell S, Hinton TV, Stevenson M. et al. Unprecedented anticancer activities of organorhenium sulfonato and carboxylato complexes against hormone-dependent MCF-7 and hormone-independent triple-negative MDA-MB-231 breast cancer cells. Mol Cell Biochem. 2018;441:151-163
64. Rakha EA, Chan S. Metastatic triple-negative breast cancer. Clin Oncol (R Coll Radiol). 2011;23:587-600
65. Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25-32
66. Aleskandarany MA, Green AR, Benhasouna AA, Barros FF, Neal K, Reis-Filho JS. et al. Prognostic value of proliferation assay in the luminal, HER2-positive, and triple-negative biologic classes of breast cancer. Breast Cancer Res. 2012;14:R3
67. Guerini-Rocco E, Piscuoglio S, Ng CK, Geyer FC, De Filippo MR, Eberle CA. et al. Microglandular adenosis associated with triple-negative breast cancer is a neoplastic lesion of triple-negative phenotype harbouring TP53 somatic mutations. J Pathol. 2016;238:677-88
68. Liu Y, Sun J, Cao W, Yang J, Lian H, Li X. et al. Dual targeting folate-conjugated hyaluronic acid polymeric micelles for paclitaxel delivery. Int J Pharm. 2011;421:160-9
69. Necela BM, Crozier JA, Andorfer CA, Lewis-Tuffin L, Kachergus JM, Geiger XJ. et al. Folate receptor-alpha (FOLR1) expression and function in triple negative tumors. PLoS One. 2015;10:e0122209
70. Walsh S, Diamond D. Non-linear curve fitting using Microsoft Excel solver. Talanta. 1995;42:561-72
71. Miao J, Tsige M, Taylor PL. Generalized model for the diffusion of solvents in glassy polymers: From Fickian to Super Case II. J Chem Phys. 2017;147:044904
72. Costa P, Sousa Lobo JM. Evaluation of mathematical models describing drug release from estradiol transdermal systems. Drug Dev Ind Pharm. 2003;29:89-97
73. Lu Y, Ding N, Yang C, Huang L, Liu J, Xiang G. Preparation and in vitro evaluation of a folate-linked liposomal curcumin formulation. J Liposome Res. 2012;22:110-9
74. Kydd J, Jadia R, Velpurisiva P, Gad A, Paliwal S, Rai P. Targeting Strategies for the Combination Treatment of Cancer Using Drug Delivery Systems. Pharmaceutics. 2017:9
75. Zhang H. Thin-Film Hydration Followed by Extrusion Method for Liposome Preparation. Methods Mol Biol. 2017;1522:17-22
76. Wang Y, Shi K, Zhang L, Hu G, Wan J, Tang J. et al. Significantly enhanced tumor cellular and lysosomal hydroxychloroquine delivery by smart liposomes for optimal autophagy inhibition and improved antitumor efficiency with liposomal doxorubicin. Autophagy. 2016;12:949-62
77. Jadia R, Scandore C, Rai P. Nanoparticles for Effective Combination Therapy of Cancer. Int J Nanotechnol Nanomed. 2016:1
78. Wang C, Wang X, Zhong T, Zhao Y, Zhang WQ, Ren W. et al. The antitumor activity of tumor-homing peptide-modified thermosensitive liposomes containing doxorubicin on MCF-7/ADR: in vitro and in vivo. Int J Nanomedicine. 2015;10:2229-48
79. Honary S, Zahir F. Effect of zeta potntial on the properties of Nano-Drug Delivery System - A Review. Tropical Journal of Pharmaceutical Research. 2013;12:265-73
80. Jadia R, Kydd J, Rai P. Remotely Phototriggered, Transferrin-Targeted Polymeric Nanoparticles for the Treatment of Breast Cancer. Photochem Photobiol. 2018;94:765-74
81. Ulbrich K, Hekmatara T, Herbert E, Kreuter J. Transferrin- and transferrin-receptor-antibody-modified nanoparticles enable drug delivery across the blood-brain barrier (BBB). Eur J Pharm Biopharm. 2009;71:251-6
82. Lee RJ, Wang S, Turk MJ, Low PS. The effects of pH and intraliposomal buffer strength on the rate of liposome content release and intracellular drug delivery. Biosci Rep. 1998;18:69-78
83. Leamon CP, Cooper SR, Hardee GE. Folate-liposome-mediated antisense oligodeoxynucleotide targeting to cancer cells: evaluation in vitro and in vivo. Bioconjug Chem. 2003;14:738-47
84. Haghiralsadat F, Amoabediny G, Helder MN, Naderinezhad S, Sheikhha MH, Forouzanfar T. et al. A comprehensive mathematical model of drug release kinetics from nano-liposomes, derived from optimization studies of cationic PEGylated liposomal doxorubicin formulations for drug-gene delivery. Artif Cells Nanomed Biotechnol. 2017:1-9
85. Heurtault B, Saulnier P, Pech B, Proust JE, Benoit JP. Physico-chemical stability of colloidal lipid particles. Biomaterials. 2003;24:4283-300
86. Heurtault B, Saulnier P, Pech B, Venier-Julienne MC, Proust JE, Phan-Tan-Luu R. et al. The influence of lipid nanocapsule composition on their size distribution. Eur J Pharm Sci. 2003;18:55-61
87. Smyth TJ, Redzic JS, Graner MW, Anchordoquy TJ. Examination of the specificity of tumor cell derived exosomes with tumor cells in vitro. Biochim Biophys Acta. 2014;1838:2954-65
88. Bazak R, Houri M, El Achy S, Kamel S, Refaat T. Cancer active targeting by nanoparticles: a comprehensive review of literature. J Cancer Res Clin Oncol. 2015;141:769-84
89. Lee J, Lilly GD, Doty RC, Podsiadlo P, Kotov NA. In vitro toxicity testing of nanoparticles in 3D cell culture. Small. 2009;5:1213-21
90. Antoni D, Burckel H, Josset E, Noel G. Three-dimensional cell culture: a breakthrough in vivo. Int J Mol Sci. 2015;16:5517-27
91. Thulasidasan AKT, Retnakumari AP, Shankar M, Vijayakurup V, Anwar S, Thankachan S. et al. Folic acid conjugation improves the bioavailability and chemosensitizing efficacy of curcumin-encapsulated PLGA-PEG nanoparticles towards paclitaxel chemotherapy. Oncotarget. 2017;8:107374-89
92. Shi HS, Gao X, Li D, Zhang QW, Wang YS, Zheng Y. et al. A systemic administration of liposomal curcumin inhibits radiation pneumonitis and sensitizes lung carcinoma to radiation. Int J Nanomedicine. 2012;7:2601-11
93. Li L, Braiteh FS, Kurzrock R. Liposome-encapsulated curcumin: in vitro and in vivo effects on proliferation, apoptosis, signaling, and angiogenesis. Cancer. 2005;104:1322-31
94. Meena R, Kumar S, Kumar R, Gaharwar US, Rajamani P. PLGA-CTAB curcumin nanoparticles: Fabrication, characterization and molecular basis of anticancer activity in triple negative breast cancer cell lines (MDA-MB-231 cells). Biomed Pharmacother. 2017;94:944-54
95. Sambale F, Lavrentieva A, Stahl F, Blume C, Stiesch M, Kasper C. et al. Three dimensional spheroid cell culture for nanoparticle safety testing. J Biotechnol. 2015;205:120-9
96. Goodman TT, Ng CP, Pun SH. 3-D tissue culture systems for the evaluation and optimization of nanoparticle-based drug carriers. Bioconjug Chem. 2008;19:1951-9
97. Jones SK, Sarkar A, Feldmann DP, Hoffmann P, Merkel OM. Revisiting the value of competition assays in folate receptor-mediated drug delivery. Biomaterials. 2017;138:35-45
98. Abraham MK, Peter K, Michel T, Wendel HP, Krajewski S, Wang X. Nanoliposomes for Safe and Efficient Therapeutic mRNA Delivery: A Step Toward Nanotheranostics in Inflammatory and Cardiovascular Diseases as well as Cancer. Nanotheranostics. 2017;1:154-65
99. Bisht S, Schlesinger M, Rupp A, Schubert R, Nolting J, Wenzel J. et al. A liposomal formulation of the synthetic curcumin analog EF24 (Lipo-EF24) inhibits pancreatic cancer progression: towards future combination therapies. J Nanobiotechnology. 2016;14:57
Author contact
![]() Corresponding author: Prakash_Raiedu, Phone: (978) 934-4971
Corresponding author: Prakash_Raiedu, Phone: (978) 934-4971